Accurate automated segmentation of autophagic bodies in yeast vacuoles using cellpose 2.0
This is the link to the article- Accurate automated segmentation of autophagic bodies in yeast vacuoles using cellpose 2.0
Here is a pdf of the article with my own annotations: https://drive.google.com/file/d/141N9QN26YRZy8xAROx_VfW6gdC1vJls7/view?usp=sharing
Annotation is a very important aspect of understanding research. What I do is send the research article to a notes apps, where I highlight parts of the article by color coding. For example, you could use red to highlight words you don't understand, and yellow to highlight important data, etc.
After reading the article, it might seem dauting at first, but after you find vocab terms you recognize with knowing the definition of unknown terms, you will find the article is not that hard after all.
Here is a list of vocab terms of the article:
Autophagy definition: It is a cellular process in which the body breaks down and recycles its own damaged or unnecessary components.
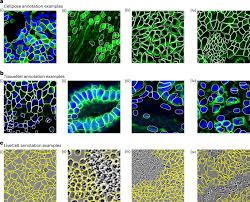
Segmentation: Division into separate parts or sections
Manual segmentation is time consuming.
Vacuolar hydrolases: Enzymes within the vacuole responsible for breaking down molecules through hydrolysis, a reaction with water
Key word: Hydrolysis - Break down of molecules by addition of water (Lysis means break)
Epoch- Training time
Protease activity: Break down proteins by cleaving peptide bonds.
What is a vector flow?
Vector flow helped find the center and edges of structures of a predefined size.
ROI: the Cellpose could learn how AB’s look like
Questions to ask when reading a research paper:
1) What is the significance of this research?
2) What is the research trying to find? (TIP: Most likely will be answered in the abstract)
3) What methods are the researchers using when conducting the research?
4) Did the research paper use references from other previous findings?
5) What was the conclusion? (TIP: The discussion section of a research paper gives this answer)
Short notes of the research:
The main aim of this research is to show how Cellpose 2.0, a computed machine, is more effective and quicker in measuring AB sizes.
Training time(epoch) should be in the median, since too much meant the the cellpose 2.0 was overcounting.
Short essay on Automated segmentation using Cell pose 2.0
Understanding “Accurate Automated Segmentation of Autophagic Bodies in Yeast Vacuoles Using Cellpose 2.0”
- Rajanya Ghosh
One of the most noticible trends in modern biology is how technology and computers now play a big role in helping us understand life on a microscopic level. A great example of this is the research article “Accurate automated segmentation of autophagic bodies in yeast vacuoles using Cellpose 2.0.” This study focused on how scientists can use artificial intelligence (AI) to identify tiny structures in cells called autophagic bodies. While that might sound complicated, the main goal was actually pretty simple: to make it faster and more accurate for researchers to study how cells clean themselves up and recycle parts using machine learning tools.
To understand this study, it helps to first know what autophagy is. “Autophagy” comes from Greek roots meaning “self-eating,” and it’s basically the process cells use to recycle their old or damaged parts. Think of it like your body’s way of taking out the trash and reusing materials to stay healthy. In yeast cells (which are often used for biological research because they share many similarities with human cells), this process happens inside an area called the vacuole. When autophagy happens, little bubbles called autophagic bodies (ABs) form and carry unwanted or broken-down materials into the vacuole to be degraded. By studying these ABs, scientists can better understand diseases in humans, since problems with autophagy are linked to things like cancer, Alzheimer’s, and other disorders.The tricky part, though, is that identifying and labeling these tiny autophagic bodies under a microscope takes a lot of time and patience. Usually, researchers have to manually go through microscope images and mark each AB, which can be both exhausting and prone to mistakes. That’s where the study’s main tool, Cellpose 2.0, comes in. Cellpose is an AI-based image analysis program that can automatically “see” and outline structures in microscope images. This is kind of like how facial recognition software can identify faces, but here it’s identifying cell structures.
The researchers in this study, including Steven K. Backues and his team, wanted to train Cellpose 2.0 to recognize autophagic bodies in yeast electron microscopy images, a very detailed type of cell photography.To do this, the team trained several AI models using different numbers of labeled autophagic bodies (called ROIs, or regions of interest). These labeled samples acted like examples for the computer—sort of like showing a student hundreds of pictures of cats so they can learn what cats look like. The researchers then tested how well each model performed using a measure called AvP (Average Precision) and IoU (Intersection over Union). These metrics tell scientists how closely the AI’s labeling matched the correct human labeling.
An IoU of 0.5 means that the AI’s and human’s outlines overlapped by 50%, while an IoU of 0.75 means they matched very closely.The results were impressive. The best model, called Model 2, performed about as well as a human expert when labeling autophagic bodies. It achieved an AvP score of 0.65 at an IoU of 0.5, which might sound modest, but it’s important to remember that even human experts only reached about 0.49 on the same test because labeling these structures is extremely hard. What’s even more interesting is that the researchers found they didn’t need thousands of training samples to get accurate results. Only around 350 labeled autophagic bodies were enough. This finding shows how efficient Cellpose 2.0 can be, making it much easier for other scientists to train their own models for different kinds of cell images.Another fascinating part of the study was how the researchers tested whether the model could handle different scenarios. They looked at yeast cells that were starved for different lengths of time (since starvation causes more autophagic activity) and even tested mutant strains of yeast where autophagic bodies were smaller. The model still performed well in these different conditions, recognizing the structures accurately. However, it struggled a bit when there were no autophagic bodies present, sometimes labeling random parts of the cell as if they were ABs. The team explained this was probably because the model hadn’t been trained on images with zero ABs.
Cellpose 2.0 showed that it could analyze images much faster than humans and with nearly the same level of accuracy.From a chemistry and biology perspective, this research is exciting because it connects molecular biology (how cells function) with computational science (how computers learn). Researchers can save tons of time analyzing data and focus more on interpreting what it all means. Instead of being stuck doing hours of manual work, researchers can now use AI to do the tedious parts while they focus on creativity and problem-solving. This project also reflects teamwork between different fieldss; all working together to answer questions about life at the smallest scale.
In conclusion, “Accurate automated segmentation of autophagic bodies in yeast vacuoles using Cellpose 2.0” is more than just a complicated-sounding study. It’s a step forward in how we use technology to explore the mysteries of biology. By teaching computers to recognize patterns in cell images, scientists can unlock new levels of understanding about how life maintains itself and fights off damage. As someone interested in science, I find it inspiring to see how research like this bridges the gap between data and discovery—reminding us that innovation doesn’t just happen in big leaps, but often in the careful, detailed work of figuring out how to see the invisible more clearly.